An Ultrafast Pre-screening Method Based on Compound Decomposition
Introduction
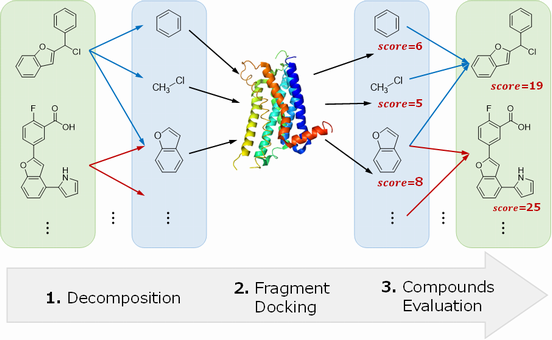
Spresso (Speedy PRE-Screening method with Segmented cOmpounds) is a novel structure-based virtual screening method based on compound decomposition. Partial structures (fragments) are often common among several compounds; therefore, the number of fragment variations neededfor evaluation is smaller than that of compounds. Our method increased calculation speeds up to approximately 200-fold compared to conventional methods.
Introduction slide (pdf)
Download
spresso_decomp_v1.0.3.tar.gz (version 1.0.3, Last update: Mar. 31 2017)
fragment_docking.py (version 1.0.2, Last update: July 8 2016)
calc_compound_score.py (version 1.0.2, Last update: July 8 2016)
Usage
Prerequisites
Codes are written in C++ and python2.
spresso_decomp
- boost (version >= 1.36)
$ sudo apt-get install libboost-all-dev $ sudo apt-get install libopenbabel-devcalc_compound_score.py
- numpy (version >= 1.10.4)
- openbabel (version == 2.4.1)
# Assumed environment is Anaconda
# numpy was already installed as a default.
$ conda install --channel https://conda.anaconda.org/Clyde_Fare openbabel Installation
spresso_decomp
$ tar -zxvf spresso_decomp_v1.0.tar.gz
$ cd spresso_decomp_v1.0
$ make -j
# If it is finished correctly, an executable file "spresso_decompose" is made.Run
1. Decomposition
$ ./spresso_decomp -c config.in
$ ./spresso_decomp -l ligandfile -f fragmentfile -o annotated_ligandfile
A sample of config.in is here.
2. Fragment Docking
$ python fragment_docking.py -g glide_protein_grid.zip -i fragment.sdf -o output.sdf -m SP3. Compounds Evaluation
$ python calc_compound_score.py annotated_ligand.sdf docked_fragment.sdf scored_ligand.sdfFor each program, "--help" option will tell you further information.
Contact
yanagisawa [at] bi.c.titech.ac.jp (Keisuke Yanagisawa, Tokyo Institute of Technology)
Reference
Keisuke Yanagisawa, Shunta Komine, Shogo D. Suzuki, Masahito Ohue, Takashi Ishida, Yutaka Akiyama. Spresso: an ultrafast compound pre-screening method based on compound decomposition, Bioinformatics, 33(23): 3836-3843, 2017. doi:10.1093/bioinformatics/btx178 [open access]