 Copyright © 2016 Akiyama Laboratory, Tokyo Institute of Technology, All Rights Reserved.
Last update: 24 Feb, 2018.
Copyright © 2016 Akiyama Laboratory, Tokyo Institute of Technology, All Rights Reserved.
Last update: 24 Feb, 2018.
|
$ cp xglide_mga.py $SCHRODINGER/mmshare-vXXXXX/python/common/
$ $SCHRODINGER/run $SCHRODINGER/mmshare-vXXXXX/python/common/xglide_mga.py inputfile.inp
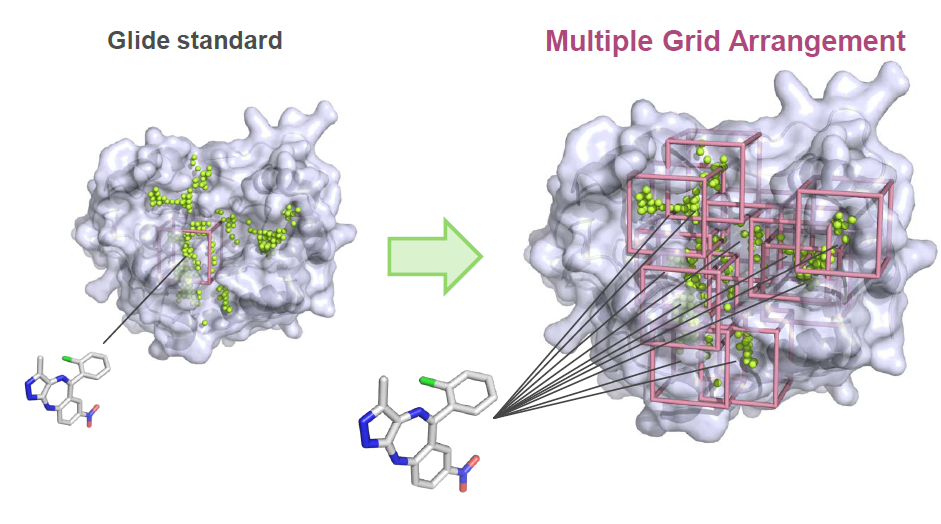
### XGlide input file ### RECEPTOR 1g9v.mae LIGAND 1g9v_ligand.mae,REFPOSE ALIGN FALSE PPREP TRUE GRIDGEN_GRID_CENTER SELF GRIDGEN_OUTERBOX 30 GRIDGEN_RANDOM_SETTING TRUE SITEMAP TRUE LIGPREP TRUE LIGPREP_EPIK TRUE DOCK_PRECISION SP DOCK_WRITE_XP_DESC FALSE DOCK_POSE_OUTTYPE poseviewer NATIVEONLY FALSE DOCK_LIG_VSCALE 0.80 SKIP_DOCKING FALSE GENERATE_TOP_COMPLEXES 1
### XGlide input file ### RECEPTOR 1g9v.mae RECEPTOR 1gkc.mae RECEPTOR 2bsm.mae LIGAND 1g9v_ligand.mae LIGAND 1gkc_ligand.mae LIGAND 2bsm_ligand.mae ALIGN FALSE PPREP TRUE GRIDGEN_GRID_CENTER SELF GRIDGEN_OUTERBOX 30 GRIDGEN_RANDOM_SETTING TRUE SITEMAP TRUE SITEMAP_MAXSITES 5 SITEMAP_FORCE TRUE LIGPREP TRUE LIGPREP_EPIK TRUE DOCK_PRECISION SP DOCK_WRITE_XP_DESC FALSE DOCK_POSE_OUTTYPE poseviewer NATIVEONLY FALSE DOCK_LIG_VSCALE 0.80 SKIP_DOCKING FALSE GENERATE_TOP_COMPLEXES 1
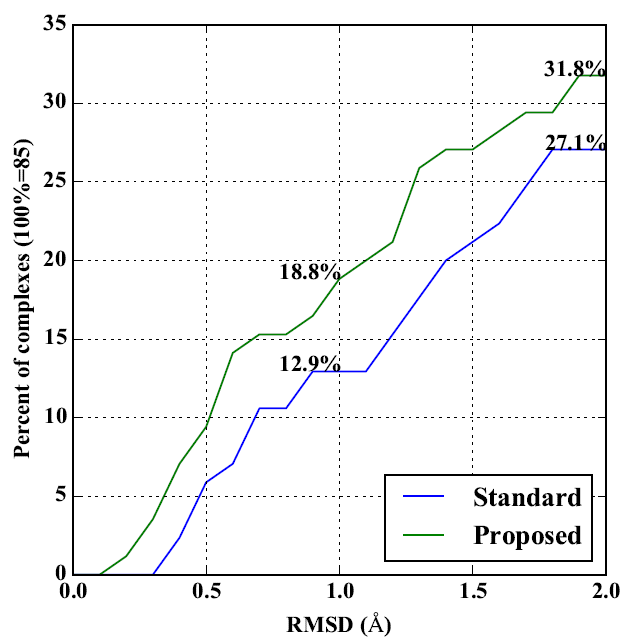
Tomohiro Ban, Masahito Ohue, Yutaka Akiyama. Multiple grid arrangement improves ligand docking with unknown binding sites: Application to the inverse docking problem, Computational Biology and Chemistry, 73: 139-146, 2018. doi: 10.1016/j.compbiolchem.2018.02.008. (open access)